Array Comparative Genomic Hybridization (aCGH)
What is Array Comparative Genomic Hybridization (aCGH)?
Array Comparative Genomic Hybridization is used to determine relative copy number variations between a test sample and reference genomic DNA. Reference or control DNA samples are fluor-labeled in one color, and the DNA of interest is labeled with another color. The two genomes are co-hybridized onto an array of DNA fragments. The relative signal intensity correlates to copy number ratio. Although, aCGH gives relative information on copy number between the genome of interest and the references genome, FISH can further be utilized to determine exact copy number in a region of interest.
CLG offers two different array comparative genomic hybridization arrays to help characterize your cell lines. CLG utilizes Agilent’s Microarray platform. Complementary FISH validation services for aCGH studies are available. (See FISH services for further information).
Services:
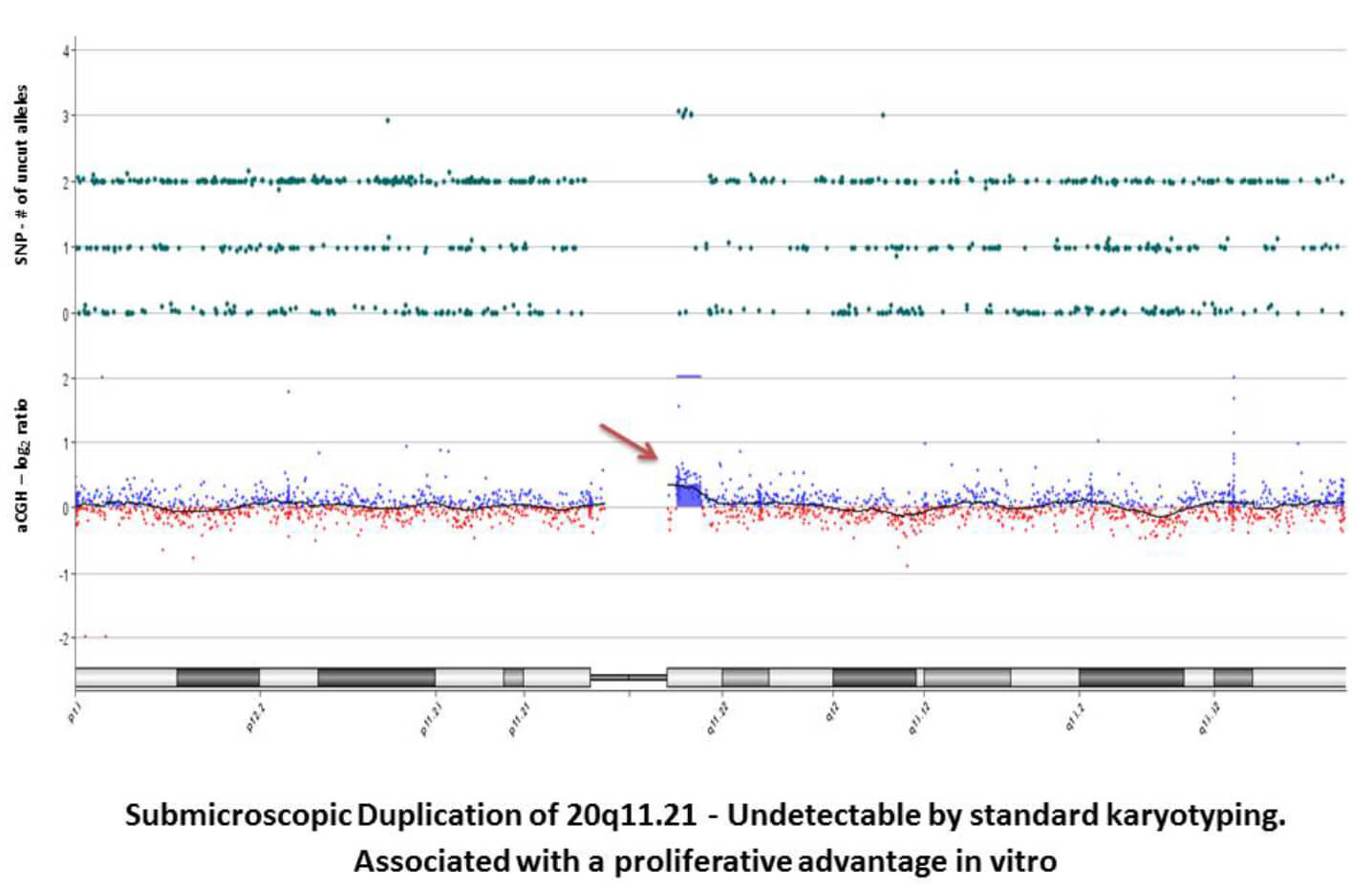
- SurePrint G3 Human CGH Microarray Kit, 8x60K: High-throughput, low-cost alternative to karyotyping. Average coverage every 41KB with increased coverage in Refseq genes (33KB)
- SurePrint G3 Human Genome CGH+SNP Microarray Kit, 4x180K: High resolution array with SNPs included. Average coverage every 25KB with increased coverage in ISCA regions (5KB)
|
SurePrint G3 Human CGH Microarray Kit, 8x60K
|
SurePrint G3 Human Genome CGH+SNP Microarray Kit, 4X180K |
|
|
| Standard FISH Services | |
| Used to: |
|
| Detects: |
|
|
Doesn’t Detect:
|
|
|
Sample Requirements:
|
|
|
Species:
|
|
|
Turnaround:
|
|
PROCEDURE: Mailing Live Cultures For FISH
Requisition Form
Each sample must be accompanied by its own Test Requisition Form. Fill out a requisition form here.
- Merz, M., Jauch, A., Hielscher, T., Mai, E. K., Seckinger, A., Hose, D., Bertsch, U., Neben, K., Raab, M. S., Salwender, H., Blau, I. W., Lindemann, H. W., Schmidt-Wolf, I., Scheid, C., Haenel, M., Weisel, K., Goldschmidt, H., & Hillengass, J. (2017). Longitudinal fluorescence in situhybridization reveals cytogenetic evolution in myeloma relapsing after autologous transplantation. Haematologica, 102(8), 1432–1438. doi.org/10.3324/haematol.2017.168005
- Shakoori A. R. (2017). Fluorescence In Situ Hybridization (FISH) and Its Applications. Chromosome Structure and Aberrations, 343–367. doi.org/10.1007/978-81-322-3673-3_16
